OMNImet•GUT | White Paper

OMNImet®•GUT enables at-home collection and ambient temperature transport of fecal samples for metabolomics
Olle M. De Bruin1, Elizaveta Freinkman2, Héloïse Breton1, Anne M. Evans2, Luke A. D. Miller2, UyenThao Nguyen2, Klaus-Peter Adam2 and Evgueni Doukhanine1
1 DNA Genotek, Ottawa, Ontario, Canada
2 Metabolon Inc, Durham, North Carolina, USA
2020-06-16
Gut metabolites derived from human and/or microbial processes, as well as via dietary intake, collectively constitute the fecal metabolome. Metabolomics can elucidate how gut metabolites affect human health and disease. Challenges surrounding the collection, stability, and transport of stool samples for metabolomics have limited the scope of studies in term of study size, accessible donor population, and geographic range available for sampling.
OMNImet™•GUT (ME-200) is a novel sampling device that provides an all-in-one solution for ambient temperature collection and transport of human fecal samples for metabolomics. Evaluation of OMNImet•GUT demonstrated that the device is suitable for at-home collection of fecal samples by untrained volunteers and is compatible with common laboratory methodologies for both untargeted and targeted metabolomics.
Furthermore, our results show that OMNImet•GUT captures an accurate and precise snapshot of fecal metabolomic profiles. Metabolomic profiles comprising >800 biochemicals per donor, including short-chain fatty acids (SCFAs), were stable for 7 days at room temperature (20°C to 26°C / 68°F to 79°F) and during simulated shipping conditions (-20°C to 30°C / -4°F to 86°F). By overcoming challenges associated with at-home collection and transport for fecal metabolomics, OMNImet•GUT will enable larger and better-controlled studies leading to key insights such as the discovery of biomarkers and drug targets.
Introduction
Metabolomics is the comprehensive measurement of hundreds to thousands of small-molecule biochemicals, or metabolites1. Metabolomics complements other ‘omics analyses including genomics, transcriptomics and proteomics by providing an integrative phenotypic readout. In the context of the gut microbiome, metabolites play a well-established role in host-microbe communication and dynamics, making metabolomics an especially powerful tool to elucidate the mechanistic functions of gut microbes in human health and disease2, 3, 4. To date, specific gut microbial metabolites have been implicated in obesity5, insulin resistance6, cardiovascular disease7, autism-spectrum disorder8 and Parkinson’s disease9, among other human health conditions. Yet, considering that most analytical methods capture only a small fraction of the biochemical diversity of gut microbiota, host physiology and their interactions, thousands of potentially pivotal molecules remain to be characterized or even discovered.
A major factor limiting the large-scale application of gut metabolomics has been the practical challenge of collecting fecal samples from human volunteers. Many metabolites in raw fecal sample are labile to rapid chemical and enzymatic degradation; therefore, immediate freezing at -80°C is recommended for preserving a sample’s metabolomic profile10 However, because feces generally cannot be produced on demand and because some donors may have difficulty traveling to a collection site, donors strongly prefer at-home collection, which precludes immediate freezing at -80°C. Currently available sampling kits for at-home collection and ambient temperature storage of fecal samples, such as DNA-stabilizing tubes and fecal immunochemical test tubes, are unsuitable for metabolomics due to high levels of detergents, buffers, salts and other additives, which are incompatible with liquid chromatography and mass spectrometry (LC/MS), the technique of choice for metabolomic analysis11, 12. Thus, there is a need for a viable collection option to enable donor-convenient, large-scale fecal metabolomics.
In response to the need for a metabolomics collection and stabilization solution, DNA Genotek™, in collaboration with Metabolon®, has developed the OMNImet•GUT at-home collection kit. This kit includes a device featuring a controlled, volumetric sample input chamber, a built-in mixing ball for rapid homogenization of the collected sample and a metabolite-stabilizing solution. In this paper, we describe the evaluation of OMNImet•GUT as an at-home collection device for ambient temperature transport of samples for fecal metabolomics analysis. Compatibility with Metabolon’s global metabolomics platform and quantitative SCFA assay is also demonstrated.
Results and Discussion
OMNImet•GUT enables at-home collection of fecal samples and integration with Metabolon’s global metabolomics platform
Without the means to stabilize metabolites at ambient temperature after at-home collection, researchers typically resort to transporting raw fecal samples on dry ice or, alternatively, freezing them in a home freezer followed by shipping on wet ice.
These approaches have major disadvantages, including the safety risks associated with study participants handling dry ice, as well as risks to metabolite stability due to temperature fluctuations during transport13 DNA Genotek already offers a unique and comprehensive solution for the collection and stabilization of microbial DNA from feces in the form of its OMNIgene®•GUT collection device.
Using the physical design of the OMNIgene•GUT kit, which ensures ease-of-use, reproducibility and homogeneity of samples14, 15, we have developed a novel complement of physical form and preservation solution for fecal metabolites, while preserving compatibility with metabolomics processing platforms, including Metabolon’s global metabolomics platform (Figure 1).
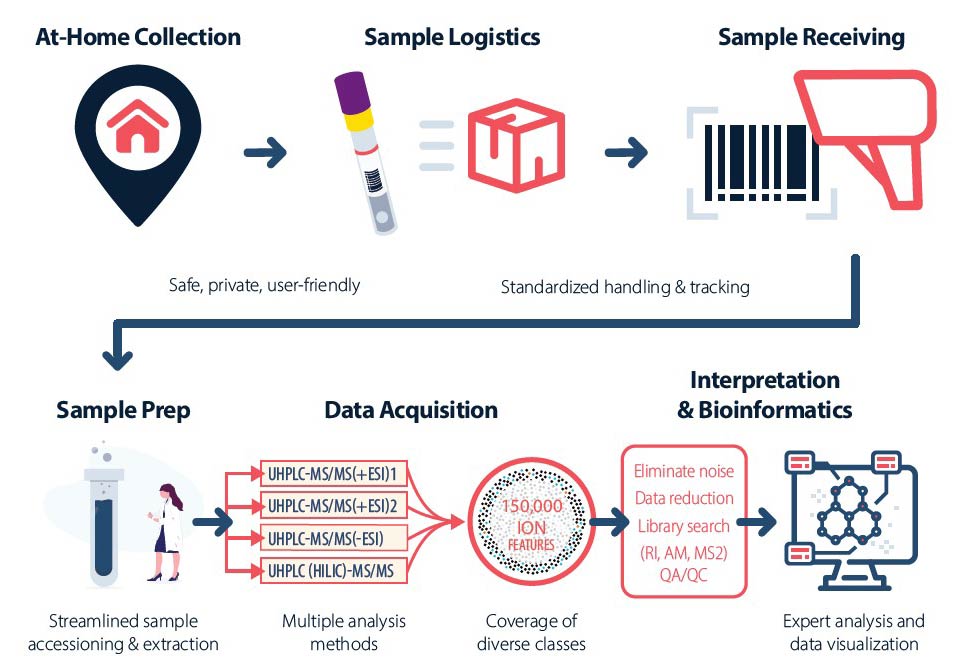
Integration with Metabolon’s global metabolomics platform.
DNA Genotek’s OMNImet•GUT
device and Metabolon’s global metabolomics platform provide an integrated solution for collection, storage,
transport and analysis of human fecal sample in metabolomics studies (figure provided by Metabolon).
Metabolite coverage is maintained after collection into OMNImet•GUT
As the gold standard of fecal sample collection, immediate freezing at -80˚C was used as the reference condition in this study. Sample collection and storage in OMNImet•GUT was compared to immediate freezing at -80˚C for sensitivity as reflected in the number of metabolites detected by Metabolon’s global metabolomics platform. Overall, the number of metabolites detected in samples from 7 donors was similar between the two collection methods (Figure 2), demonstrating that collection with OMNImet•GUT does not introduce a systematic bias or experimental artefacts and maintains a gold standard level of detection sensitivity for metabolomics analysis.
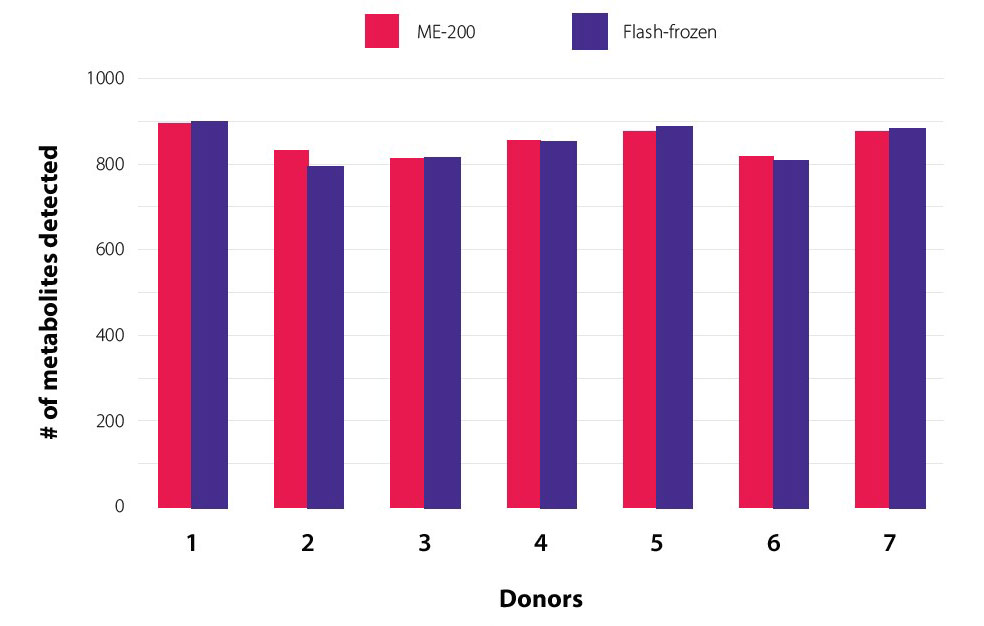
Preservation of metabolomic coverage.
Technical
replicate samples (n=3) from each of 7 donors were collected into
either a flash-frozen or an OMNImet•GUT device (ME-200), followed
by immediate freezing at -80°C. The number of metabolites detected
in all technical replicates (n=3) of each donor’s samples is plotted.
Metabolite stability and analytical precision are maintained after collection into OMNImet•GUT
To be suitable for at-home collection and storage of fecal samples, a device must not distort the metabolomic profile of the raw fecal sample and must adequately stabilize metabolites over time. To assess the fidelity and stability of metabolomic profiles from OMNImet•GUT samples, Spearman correlations were calculated for all detected metabolites between flash-frozen samples and matched OMNImet•GUT samples held for 0, 1, 4 and 7 days at room temperature (table 1).
| Condition | T0 | T1 | T4 | T7 | Freeze-thaw |
|---|---|---|---|---|---|
| Median R | 0.89 | 0.88 | 0.86 | 0.86 | 0.89 |
Table 1: Correlation analysis across n=7 biological replicates (after averaging each donor’s n=3 technical replicates) in each OMNImet•GUT storage condition (baseline (T0), room temperature for 1, 4, or 7 days (T1, T4, T7), or freeze-thaw) vs. flash-frozen. The median Spearman coefficient across all detected metabolites is shown.
The median Spearman coefficient between flash-frozen and OMNImet•GUT-collected samples held at room temperature for 0 days (i.e., samples immediately frozen in OMNImet•GUT devices) was 0.89, indicating that OMNImet•GUT preserves the metabolomic profile of the raw sample with a high degree of fidelity. Furthermore, median Spearman correlations to flash-frozen remained in the 0.86-0.88 range at 1, 4 and 7 days of room temperature storage in OMNImet•GUT, indicating sustained metabolite stabilization within the device.
Further analysis of Spearman coefficients after grouping metabolites by their biochemical classification (superpathway) revealed that peptides were likely a contributing factor of discrepancy between flash-frozen and OMNImet•GUT samples at all timepoints (Figure 3). The 37 peptide superpathway biochemicals detected in this study were primarily dipeptides, which may originate from food sources, gut tissue and/or microbial activity and whose importance in fecal metabolomics is currently unclear. By contrast, the amino acid and xenobiotic superpathways, which contain most of the known bioactive microbial metabolites, displayed median Spearman coefficients in the 0.9 range at all timepoints (T0 to T7) in OMNImet•GUT relative to matched flash-frozen samples (Figure 3). These results indicate that storage in OMNImet•GUT preserves the vast majority of metabolites in raw fecal sample with both fidelity and stability.
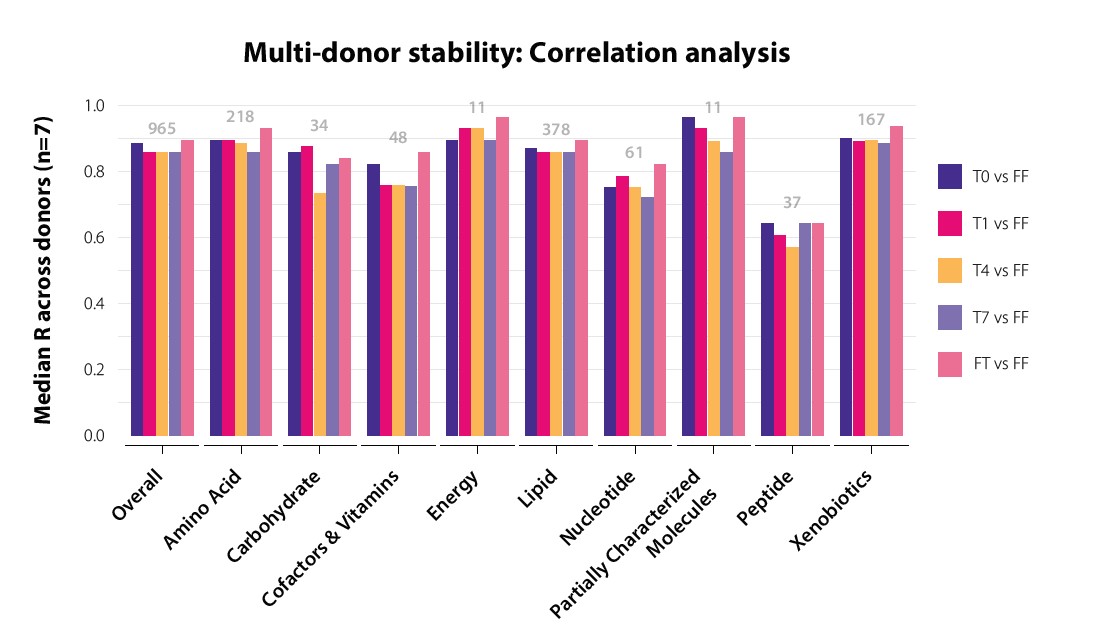
Accurate measurement of individual metabolites and
metabolic pathways.
Correlation analysis across n=7 biological
replicates (after averaging each donor’s n=3 technical replicates) in
each OMNImet•GUT storage condition (baseline (T0), room temperature
for 1, 4, or 7 days (T1, T4, T7), or freeze-thaw) vs. flash-frozen (FF).
The median Spearman coefficient across each major class of biochemicals
(metabolic superpathway) is plotted and the number of biochemicals
in each superpathway (detected in flash-frozen samples) is labeled.
Because metabolite stabilization is such a critical performance metric for an at-home fecal collection and storage device, we sought to obtain another quantitative measure of metabolite stability over time in OMNImet•GUT. In this case, T0 samples (i.e., samples immediately frozen in OMNImet•GUT devices) were used as the reference condition in order to isolate the time effect. Intraclass Correlation Coefficients (ICC) between T0 and subsequent timepoints were calculated as described12, using samples from 7 individual donors. The median ICC across all detected metabolites after 4 and 7 days of room temperature storage in OMNImet•GUT was 0.93 and 0.90, respectively (maximum = 1.0). ICC values above 0.9 reflect excellent agreement between datasets16,17 , again indicating successful stabilization of metabolites over 7 days of storage at room temperature.
Given the relatively minor discrepancies between flashfrozen samples and samples stored in OMNImet•GUT for up to 7 days, we sought to assess the impact of storing samples in OMNImet•GUT on the ability to derive biological insights from human subject data. To do this, we performed Hierarchical Clustering Analysis (HCA) and Principal Component Analysis (PCA) on matched samples from 7 donors that had been flashfrozen or stored in OMNImet•GUT for varying lengths of time. In both analyses, samples clustered clearly by donor, while differences among storage conditions and timepoints were minor relative to inter-donor differences (Figures 4 and 5).
In particular, no timedependent trends were consistently observed in either the PCA or HCA. These results indicate that storage in OMNImet•GUT for up to 7 days at room temperature preserves the unique metabolomic fingerprint of human fecal samples. Taken together, the Spearman, ICC, PCA and HCA analyses demonstrate that OMNImet•GUT samples continue to accurately reflect the metabolomic profile of raw fecal samples after storage for up to 7 days at room temperature.
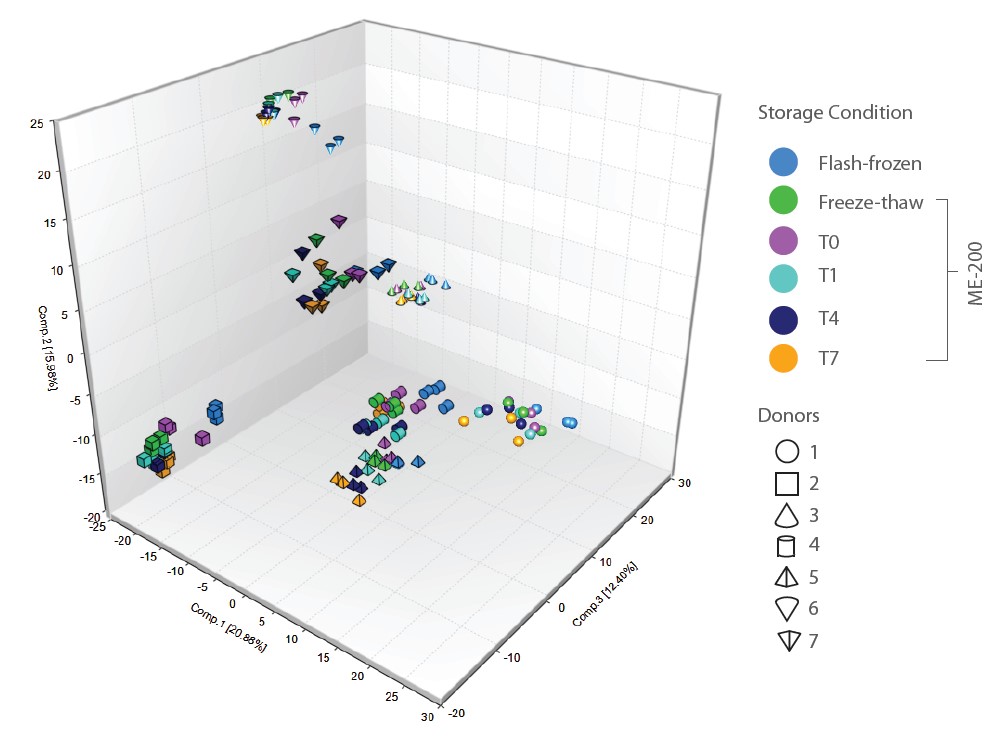
Figure 4: PCA. Principal component analysis of metabolomic data collected using fecal samples from 7 donors (n=3 technical replicates per donor) that were flash-frozen or stored in OMNImet•GUT as indicated: baseline (T0), room temperature for 1, 4, or 7 days (T1, T4, T7), or freeze-thaw.
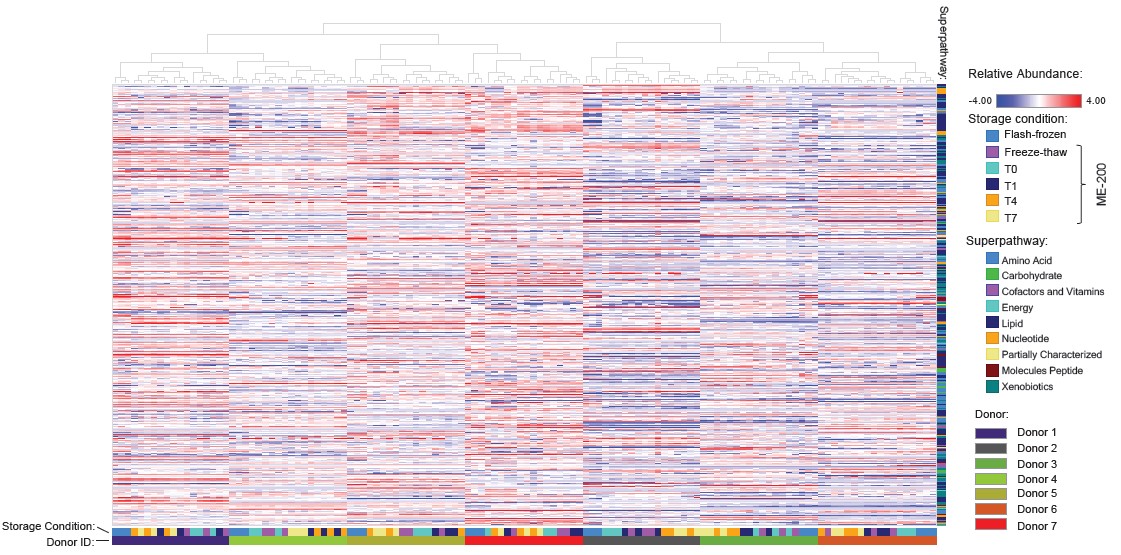
Figure 5: HCA. Hierarchical clustering analysis of metabolomics data collected using fecal samples from 7 donors (n=3 technical replicates per donor) that were flash-frozen or stored in OMNImet.GUT as indicated: baseline (T0), room temperature for 1, 4, or 7 days (T1, T4, T7), or freeze-thaw. (The same dataset as Figure 4).
In addition to stability, precision is a critical consideration in metabolomic analysis. Therefore, we sought to benchmark the precision of OMNImet•GUT samples against that of flash-frozen samples. PCA (Figure 4) showed that within-donor variability (n=3 technical replicates per condition from each of 7 donors) was comparable for samples in OMNImet•GUT and flash-frozen samples. In a separate analysis of n=6 technical replicates per condition collected from each of three donors, no statistically significant differences were observed between the median relative standard deviation (RSD) across all biochemicals in the OMNImet•GUT samples relative to matched flash-frozen replicates (data not shown). Together, these data indicate that collection into OMNImet•GUT device does not introduce additional technical variability into metabolomic analyses relative to the gold standard collection method.
OMNImet•GUT is compatible with freezethawing and simulated shipping conditions
Simulating transport of samples through different temperature conditions was performed by subjecting samples collected into OMNImet•GUT to freezethaw at -20˚C followed by 30˚C. This treatment simulates a scenario of air transport followed by ground transport without temperature control, or, alternatively, samples that have been frozen (e.g., in a household freezer) and then delivered to a study site without temperature control.
As seen through PCA, HCA and Spearman correlation (Figures 4, 5 and table 1), freeze-thaw cycling had minimal effect on the metabolomic profile of samples from all donors, indicating that OMNImet•GUT-collected samples are not sensitive to freeze-thawing and can be shipped without temperature-controlled conditions.
Short-chain fatty acids are stable in OMNImet•GUT for at least 7 days at room temperature
Short-chain fatty acids (SCFAs) are key mediators of gut microbial activity and play important roles in regulating the immune system, maintaining the gut epithelial barrier and preventing disorders such as metabolic syndrome, inflammatory bowel disease and certain types of cancer18. Therefore, we assessed the performance of OMNImet•GUT in stabilizing SCFAs at room temperature. To this end, a pooled fecal sample derived from 7 donors was stored at room temperature for varying lengths of time in OMNImet•GUT and in an unstabilized condition (cryovial), followed by targeted, quantitative analysis of eight SCFAs. While concentrations of SCFAs in raw samples increased approximately 2- to 10-fold after 96h (4 days) at room temperature, concentrations in OMNImet•GUT samples remained unchanged between 0 and 13 days at room temperature (Figure 6). These results indicate that OMNImet•GUT quenches bacterial metabolism and that SCFAs are stable in the device for at least 7 days at room temperature. In a similar experiment with samples from 7 individual donors, all SCFAs from all donors remained stable in OMNImet•GUT for 7 days at room temperature (data not shown). These results demonstrate that OMNImet•GUT is compatible with Metabolon’s short-chain fatty acid assay and stabilizes SCFAs for at least 7 days at room temperature.
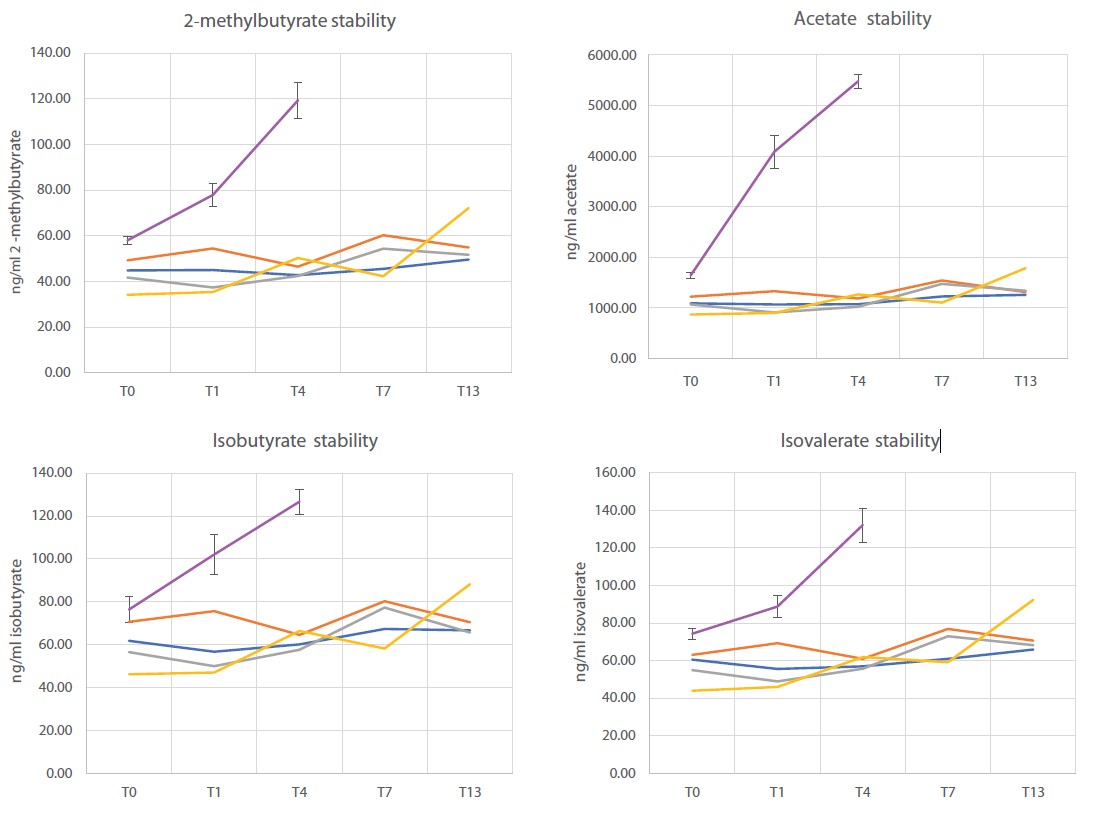
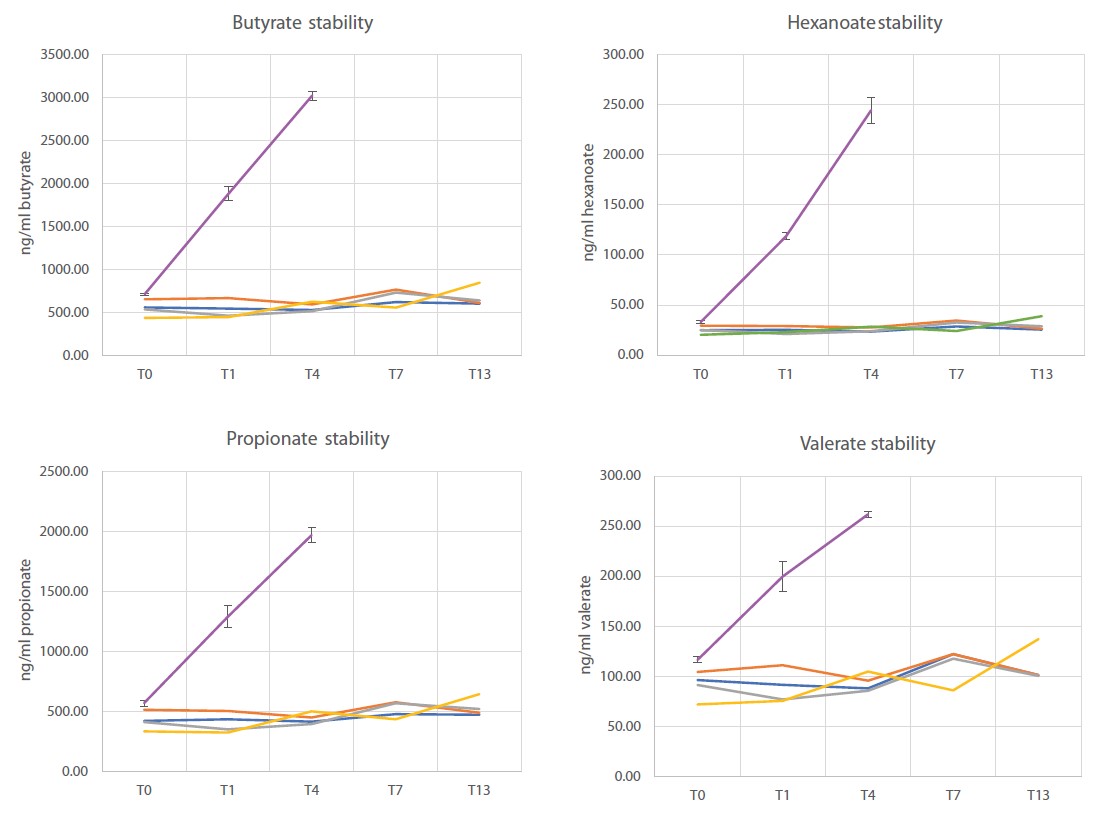

Stability of SCFA in OMNImet•GUT
Aliquots of a 7-donor pooled raw fecal sample were either left unstabilized (n=4 replicates) or
stored in OMNImet•GUT (ME-200) (n=4 replicates, A/B/C/D) at room temperature for the indicated number of days (T0 to T13), followed by
quantification of eight short-chain fatty acids (SCFAs) using targeted LC-MS/MS.
Conclusions
In the context of most fecal metabolomics studies, at-home collection is necessary to recruit a representative donor population and achieve adequate statistical power, which are prerequisites for identification of meaningful and translatable metabolite-disease associations. To date, no standardized, validated at-home collection kit for fecal metabolomics has been available.
Measurement of >800 individual biochemicals per human donor on Metabolon’s global metabolomics platform, as well as quantitative analysis of 8 SCFAs, showed that samples collected in OMNImet•GUT devices remained stable at room temperature for up to 7 days and recapitulated the metabolomic profiles of matched flash-frozen samples while maintaining similar technical precision. These results show that sample collection and storage in OMNImet•GUT compares favorably with the gold standard flashfreezing methodology. OMNImet•GUT will enable the collection of high-quality fecal metabolomics data at a scale previously not possible and drive a new wave of discoveries in the field.
For Materials and methods: OMNImet•GUT (ME-200) enables at-home collection and ambient temperature transport of fecal samples for metabolomics ( pdf )
References
1 Schrimpe-Rutledge, A.C., Codreanu, S.G., Sherrod, S.D., and McLean, J.A. (2016). Untargeted Metabolomics Strategies-Challenges and Emerging
Directions. J Am Soc Mass Spectrom 27, 1897-1905. http://doi.org/10.1007/s13361-016-1469-y
2 Lamichhane, S., Sen, P., Dickens, A.M., Oresic, M., and Bertram, H.C. (2018). Gut metabolome meets microbiome: A methodological perspective
to understand the relationship between host and microbe. Methods 149, 3-12. http://doi.org/10.1016/j.ymeth.2018.04.029
3 Smirnov, K.S., Maier, T.V., Walker, A., Heinzmann, S.S., Forcisi, S., Martinez, I., Walter, J., and Schmitt-Kopplin, P. (2016). Challenges of metabolomics
in human gut microbiota research. Int J Med Microbiol 306, 266-279. http://doi.org/10.1016/j.ijmm.2016.03.006
4 Lee-Sarwar, K.A., Lasky-Su, J., Kelly, R.S., Litonjua, A.A., and Weiss, S.T. (2020). Metabolome-Microbiome Crosstalk and Human Disease. Metabolites 10.
http://doi.org/10.3390/metabo10050181
5 Liu, R., Hong, J., Xu, X., Feng, Q., Zhang, D., Gu, Y., Shi, J., Zhao, S., Liu, W., Wang, X., et al. (2017). Gut microbiome and serum metabolome alterations
in obesity and after weight-loss intervention. Nat Med 23, 859-868. http://doi.org/10.1038/nm.4358
6 Pedersen, H.K., Gudmundsdottir, V., Nielsen, H.B., Hyotylainen, T., Nielsen, T., Jensen, B.A., Forslund, K., Hildebrand, F., Prifti, E., Falony, G., et al. (2016).
Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376-381. http://doi.org/10.1038/nature18646
7 Wang, Z., Klipfell, E., Bennett, B.J., Koeth, R., Levison, B.S., Dugar, B., Feldstein, A.E., Britt, E.B., Fu, X., Chung, Y.M., et al. (2011). Gut flora metabolism
of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57-63. http://doi.org/10.1038/nature09922
8 Sharon, G., Cruz, N.J., Kang, D.W., Gandal, M.J., Wang, B., Kim, Y.M., Zink, E.M., Casey, C.P., Taylor, B.C., Lane, C.J., et al. (2019). Human Gut Microbiota
from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 177, 1600-1618 e1617. http://doi.org/10.1016/j.cell.2019.05.004
9 Hertel, J., Harms, A.C., Heinken, A., Baldini, F., Thinnes, C.C., Glaab, E., Vasco, D.A., Pietzner, M., Stewart, I.D., Wareham, N.J., et al. (2019). Integrated
Analyses of Microbiome and Longitudinal Metabolome Data Reveal Microbial-Host Interactions on Sulfur Metabolism in Parkinson’s Disease. Cell Rep 29,
1767-1777 e1768. http://doi.org/10.1016/j.celrep.2019.10.035
10 Stevens, V.L., Hoover, E., Wang, Y., and Zanetti, K.A. (2019). Pre-Analytical Factors that Affect Metabolite Stability in Human Urine, Plasma, and Serum:
A Review. Metabolites 9. http://doi.org/10.3390/metabo9080156
11 Lim, M.Y., Hong, S., Kim, B.M., Ahn, Y., Kim, H.J., and Nam, Y.D. (2020). Changes in microbiome and metabolomic profiles of fecal samples stored with
stabilizing solution at room temperature: a pilot study. Sci Rep 10, 1789. http://doi.org/10.1038/s41598-020-58719-8
12 Loftfield, E., Vogtmann, E., Sampson, J.N., Moore, S.C., Nelson, H., Knight, R., Chia, N., and Sinha, R. (2016). Comparison of Collection Methods for Fecal
Samples for Discovery Metabolomics in Epidemiologic Studies. Cancer Epidemiol Biomarkers Prev 25, 1483-1490. http://doi.org/10.1158/1055-9965.EPI-16-0409
13 Gratton, J., Phetcharaburanin, J., Mullish, B.H., Williams, H.R., Thursz, M., Nicholson, J.K., Holmes, E., Marchesi, J.R., and Li, J.V. (2016). Optimized Sample
Handling Strategy for Metabolic Profiling of Human Feces. Analytical Chemistry 2016 88 (9), 4661-4668. http://doi.org/10.1021/acs.analchem.5b04159
14 Doukhanine, E., Bouevitch, A., Pozza, L., and Merino, C. (2014). OMNIgene®•GUT enables reliable collection of high quality fecal samples for gut
microbiome studies. PD-WP-00040, DNA Genotek, Inc. Available at https://dnagenotek.com/US/pdf/PD-WP-00040.pdf.
15 Doukhanine, E., Egloff, C., Bouevitch A., Lynch D., Smith C., and Iwasiow R. (2016). Choice of microbiome extraction methodology significantly affects
biomarker discovery sensitivity and requirements for cohort size. MK-00670, DNA Genotek, Inc.
16 Cicchetti, D. V. (1994). Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology.
Psychological Assessment, 6(4), 284–290. https://doi.org/10.1037/1040-3590.6.4.284
17 Koo, T.K., and Li, M.Y. (2016). A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med 15,
155-163. http://doi.org/10.1016/j.jcm.2016.02.012
18 Tan, J., McKenzie, C., Potamitis, M., Thorburn A.N., Mackay C.R., and Macia, L. (2014). The role of short-chain fatty acids in health and disease. Adv
Immunol, 121:91‐119. doi:10.1016/B978-0-12-800100-4.00003-9
Beyond GUT microbiome
The OMNIgene® line of products also includes all-in-one systems for quantitative profile analysis of the oral and vaginal microbiome.
View All Microbiome Collection DevicesInside DNA Genotek
Legal