- Costea PI et al. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol. Nov;35(11):1069-1076 (2017).
- OMNIgene•GUT enables reliable collection of high quality fecal samples for gut microbiome studies. DNA Genotek. PD-WP-00040. (2016).
- Yu Z and Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques. May;36(5):808-12. (2004).
- Song EJ, Lee ES and Nam YD. Progress of analytical tools and techniques for human gut microbiome research. J Microbiol. Oct;56(10):693-705. (2018).
- Choo JM, Leong L and Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep. 2015 Nov 17;5:16350. (2015).
Evaluation of DNA extraction methods to obtain accurate and reliable results from gut microbiome samples
Evaluation of DNA extraction methods to obtain accurate and reliable results from gut microbiome samples
B. LeFrançois and L. Cunningham
DNA Genotek, Ottawa, Ontario, Canada
2019-01-10
Introduction
When it comes to microbial community profiling from complex environments like stool, sources of technical variation such as collection procedures, storage conditions and sample extraction can greatly impact the accuracy of the data. In conjunction with sample stabilization buffer offering post-collection protection from microbial profile changes, the choice of extraction methodology for obtaining sufficient and high quality microbial nucleic acids is essential for meaningful and reliable insights into these complex communities.
Despite the lack of a standardized or optimized extraction method for efficiently recovering DNA from all stool sample types, most methods developed over the years rely on mechanical and/or enzymatic lysis of microbial cells to release DNA. The lysis component of the extraction method plays a critical role in the efficiency of DNA recovery, especially for challenging sample matrices (presence of fibers or undigested particles) or low biomass samples (ex., infant or donors undergoing dysbiosis). A recent comparison of extraction protocols showed that mechanical lysis (i.e., bead beating) is paramount for efficient DNA release from tough to lyse gram-positive bacteria and is positively correlated with increased sample diversity.1
DNA Genotek’s OMNIgene•GUT collection and stabilization devices constitute a reliable method to easily and accurately capture the microbial community present in stool samples at the time of collection.2 In this application note, we tested and compared the extraction performance of four commonly used bead beating based DNA extraction methods on stool samples collected with the OMNIgene•GUT device.
Methods
Stool samples from eight adult human donors were collected in OMNIgene•GUT under IRB guidance and DNA was extracted from a 250 µL aliquot using either the QIAamp® PowerFecal® DNA Kit (QIAGEN®), the QIAamp PowerFecal Pro DNA Kit (QIAGEN), the RBB+C method3 or the ZymoBIOMICS™ DNA Miniprep Kit (Zymo Research). Extractions were performed within 72 hours of sample collection into OMNIgene•GUT. DNA yields, lysis efficacy as well as the quality and integrity of the recovered DNA was compared for these four methods.
For the PowerFecal (QIAamp PowerFecal DNA Kit) and PowerFecal Pro (QIAamp PowerFecal Pro DNA Kit) extractions, bead beating was performed for 10 minutes using a 2 mL tube vortex adapter (QIAGEN). For the RBB+C extractions, bead beating was performed on a Bead Ruptor Elite (OMNI International) at 5.5 m/s for 3 minutes, conditions that were found to be optimal for lysis while maintaining good DNA integrity. For ZymoBIOMICS (ZymoBIOMICS DNA Miniprep Kit) extractions, bead beating was also performed on the Bead Ruptor Elite using conditions recommended by Zymo Research technical support (5 cycles consisting of 1 minute bead beating at 6.5 m/s with 5 minutes incubation on ice between each cycle).
Results
Our extraction data demonstrate that PowerFecal Pro produced the highest average DNA yield across samples collected from multiple donors, followed by ZymoBIOMICS, RBB+C and PowerFecal (Figure 1). Interestingly, RBB+C showed the least variability from donor to donor while the performance of ZymoBIOMICS appears to be highly donor dependent.
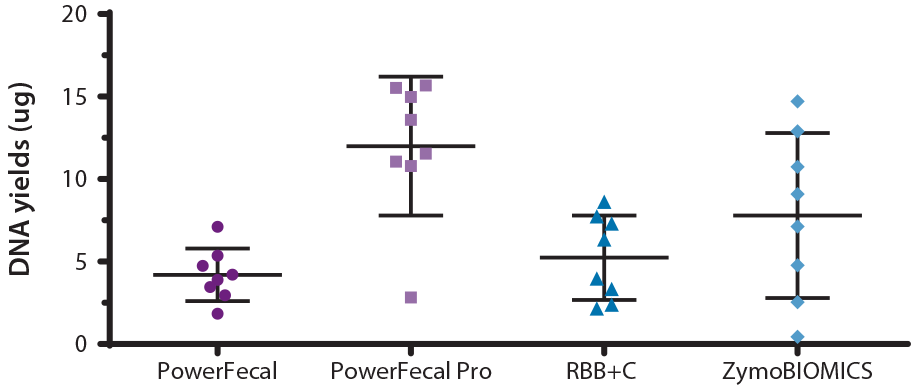
Figure 1: Total DNA yields following extraction of a 250 µL OMNIgene•GUT aliquot (representing ~50 mg of fecal material) from 8 different donors using either PowerFecal, PowerFecal Pro, RBB+C or ZymoBIOMICS. DNA Extractions were performed as per manufacturer’s instructions. The eluted DNA was quantified using the PicoGreen® assay.
To assess lysis efficiency of the various extractions, we quantified the relative levels of two hard to lyse gram-positive bacteria (Bifidobacterium longum and Faecalibacterium prausnitzii) and gram-negative bacteria (Bacteroides genus) in 10 ng total DNA using real-time PCR.
Lysis of the gram-positive bacteria was suboptimal in PowerFecal extracted samples (Figure 2), as demonstrated by the significantly higher Ct values for Bifidobacterium longum and Faecalibacterium prausnitzii. A higher Ct value is correlated with lower levels of the bacterial DNA in the sample and indicates suboptimal lysis of the assayed species. In contrast, PowerFecal Pro, RBB+C and ZymoBIOMICS extractions all gave lower Ct values, indicative of a more efficient lysis. As expected, levels of the easy to lyse gram-negative bacteria genus Bacteroides were similar across extractions.
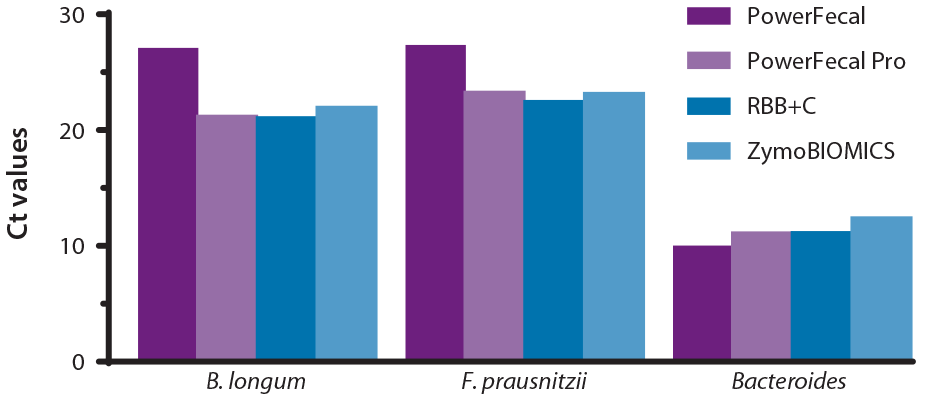
Figure 2: Relative levels of gram-positive or negative bacteria in stool DNA samples extracted using PowerFecal , PowerFecal Pro , RBB+C or ZymoBIOMICS. Levels of Bifidobacterium longum (gram+), Faecalibacterium prausnitzii (gram+) or Bacteroides species (gram- genus) were measured in real-time PCR assays using specific primer pairs and 10ng total DNA as template. Data presented here is from a representative donor.
To evaluate the quality and integrity of the extracted DNA, we checked samples for the presence of PCR inhibitors and measured DIN values (DNA Integrity Number). Fecal carry-over contaminants, such as humic acids and polyphenols, can impact downstream processing and analysis of the samples by interfering with PCR and DNA library preparation for next generation sequencing platforms. All DNA samples extracted with commercially available kits were free of inhibitors, but we were able to detect inhibitors in 10-20% of RBB+C extracted samples (data not shown). Furthermore, our results demonstrate that PowerFecal, PowerFecal Pro and RBB+C all produced high quality, high molecular DNA with an average fragment size of >10 kbp and DIN values higher than 7. (Figure 3 and Table 1). In comparison, ZymoBIOMICS extractions yielded significantly sheared DNA with an average fragment size of 3–4 kbp and average DIN values lower than 5.
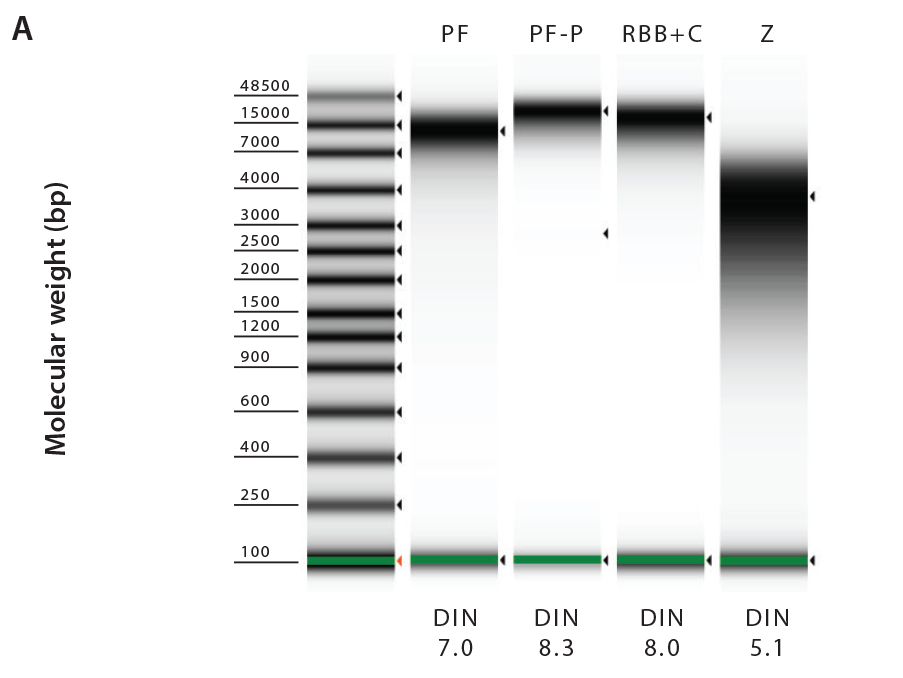
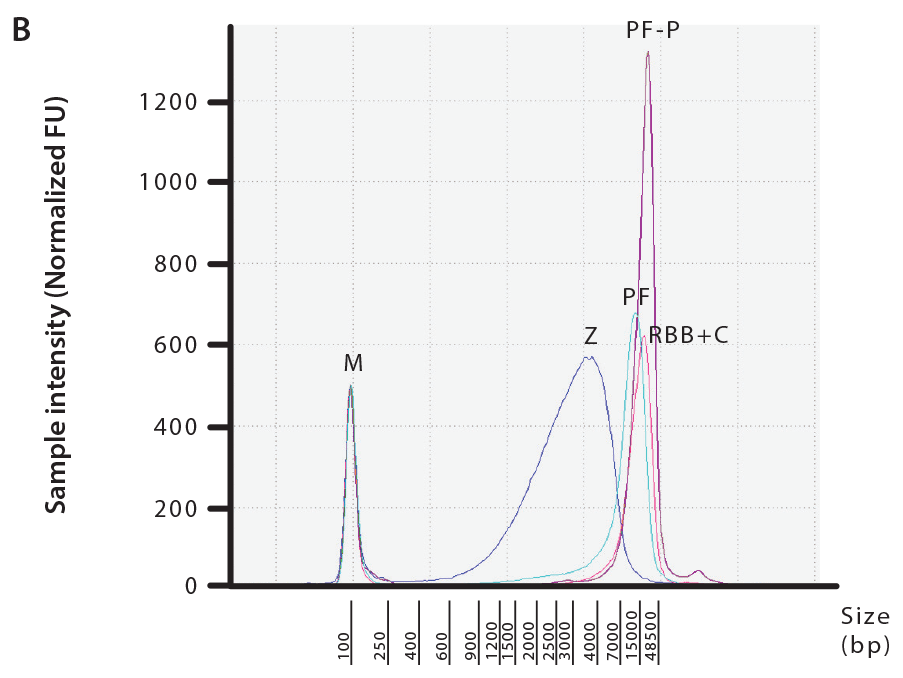
Figure 3: Integrity and average size of the DNA samples extracted from OMNIgene•GUT samples using PowerFecal (PF), PowerFecal Pro (PF-P), RBB+C or ZymoBIOMICS (Z) (M = Control Marker). Extracted DNA samples were run on a genomic DNA ScreenTape (Agilent) to determine DNA integrity (A) and average fragment size (B). Data presented here is from a representative donor.
PowerFecal |
PowerFecal Pro |
RBB+C |
ZymoBIOMICS |
|
DNA yields |
Low to medium |
Medium to high |
Low to medium |
Medium to high |
Lysis efficacy |
Limited on gram + bacteria |
Good |
Good |
Good |
Average fragment size Average DIN |
19.2 kbp ± 11.5 7.1 ± 1.0 |
19.3 kbp ± 2.3 7.8 ± 0.6 |
18.6 kbp ± 1.4 7.9 ± 0.3 |
3.2 kbp ± 0.6 4.8 ± 0.5 |
Presence of PCR/library prep inhibitors |
Absent |
Absent |
Detectable in
10-20% of samples |
Absent |
Table 1: Summary of the DNA yields, lysis efficacy, DNA quality and presence of PCR inhibitors obtained following DNA extraction from OMNIgene•GUT stool samples (n=8) using various DNA extraction kits: QIAamp PowerFecal DNA Kit, the QIAamp PowerFecal Pro DNA Kit, the RBB+C protocol or the ZymoBIOMICS DNA Miniprep Kit.
It is worth noting that the performance of the extraction kits on fresh fecal samples was similar in every aspect to OMNIgene•GUT collected samples (data not shown). This indicates that DNA Genotek’s stabilizing chemistry has no impact on the overall performance of the DNA extraction kits tested in this application note.
Conclusions
Choosing a reliable stool DNA extraction protocol is critical to any microbiome study. Efficient and consistent lysis of the highly diverse microbial species present in stool samples is a key consideration to ensure that the extracted DNA captures an accurate representation of the bacterial community present in the sample with minimal bias. Suboptimal lysis, such as seen with the PowerFecal kit, can significantly impact the measured relative abundance of gram-positive species such as Faecalibacterium prausnitzii and Bifidobacterium longum. Interestingly, several studies have reported a bias in F. prausnitzii4,5 in OMNIgene•GUT collected samples. Multiple lines of evidence indicate that these findings are derived from suboptimal DNA extractions rather than actual changes in the sample taxonomic composition upon collection and stabilization of the samples.
Isolation of high molecular weight DNA is a requirement to take advantage of the full suite of available next generation sequencing technologies. Long range sequencing applications such as full-length 16S or Nanopore based sequencing, are dependent on the recovery of relatively intact and high quality DNA. Our data suggests that PowerFecal Pro and RBB+C extractions provide a good balance between lysis efficiency while preserving DNA integrity, while the ZymoBIOMICS lysis is achieved at the expense of DNA integrity. This result has also been observed in preliminary testing of Zymo Research Quick-DNA™ Fecal/Soil Microbe Kit.
In summary, out of the extraction methods evaluated, we have identified PowerFecal Pro as the best performing stool DNA extraction kit, giving (i) high DNA yields from a wide range of donors, (ii) efficient lysis of gram+ bacteria, and (iii) recovery of high molecular weight DNA free of inhibitors. Moreover, our preliminary data also indicates that PowerFecal Pro performs well with low biomass samples (such as infant and donors undergoing dysbiosis), making this extraction method a good choice for a wide variety of gut microbiome studies. We highly recommend using the OMNIgene•GUT collection and stabilization device to effectively capture an accurate representation of the in vivo state of the donor microbial profile, without introducing bias or loss of DNA integrity. For extracting OMNIgene•GUT samples, we recommend the PowerFecal Pro extraction kit in order to minimize any potential extraction bias. This combination will provide the most accurate and thorough representation of the microbial community present at the time of sample collection.
References
OMNIgene®•GUT (OM-200) is not available for sale in the United States.
OMNIgene®•GUT (OMR-200) is for research use only, not for use in diagnostic procedures.
Some DNA Genotek products may not be available in all geographic regions.
®OMNIgene is a registered trademark of DNA Genotek Inc. All other brands and names contained herein are the property of their respective owners.
All DNA Genotek protocols, white papers and application notes, are available in the support section of our website at www.dnagenotek.com.
Inside DNA Genotek
Legal


